Compound queries are used for collecting and returning data. This can be accomplished by returning results from multiple queries; aggregating data; getting counts, averages and lists; working with search server sets; and performing advanced analytics and creating charts.
Print()
To view the results of the script, use the print function. Print will display the results in the output tab found to the right of the script area of Report Builder.
print(MyVariable)It is important to note that there is a functionality difference in the use of print and the use of return when writing queries. While print makes the results available for the user to see, return makes the results available for use in other functions of the script when referencing the return to the response object.
Monospace Font
By default the results printed to the output is not in a monospace font. However, if text alignment is necessary for readability, add a work-around that allows the user to change the response to a monospace font. To do so, add <code></code> tags around the string formatting in the print function.
column1 = "name"
column2 = "lab result"
data = {"bob":5.789765,
"sue":3.235789,
"john":7.458915,
"alice":1.689066}
print('<code>%-7s\t%s</code>' % (column1, column2)) # the - indicates left alignment (right align by leaving off the -), the column length is 7 characters, \t adds a tab
for name, value in data.items():
print('<code>%-7s\t%1.2f</code>' % (name, value))The response from the above query would look something like this:
name lab result
bob 5.78
sue 3.23
john 7.45
alice 1.68Wrapper Functions
IMAT Solutions has created some wrapper or helper, functions that increase the efficiency of report writing. For instance, if the report writer wants to find all unique values within a certain field, using get_distinct_values() can accomplish this goal in one line of code; whereas, it would take multiple lines without the wrapper function. Wrapper functions also lessen the likelihood of errors occurring in the query. Each function has specific parameters that can be used within the function.
- get_hits
- get_facets
- show_hits
- get_distinct_values
- get_unique_values
- get_hits_total
- get_result_string
- output_plot
get_hits()
get_hits(q, fields, store=None, limit=-1, outputset=None, advanced_options=None)
The use of get_hits() replaces the previous Query method and allows the server to return a list of hits that match the query criteria. Assign the query a name and begin each query with get_hits. Follow it with a Boolean expression and parameters.
Six parameters can be used with get_hits(): q, fields, store, limit, outputset, advanced_options. See the Query Syntax guide for details on each of these parameters.
query_name = get_hits('()diabetes', store=2, fields=CLINICAL_REPORTS_FIELDS, limit=10, outputset=True, advanced_options={'dtype':{'extract.patient_id': numpy.object_}})get_facets()
get_facets(q, facets=True, store=0, countby=None)
To use get_facets(), assign the query a name, begin the query with get_facets, then follow it with a Boolean expression and the facet and store parameters. This function requires these two parameters in order to operate: q and facets. The facets parameter must include the name of the field followed by a colon and the word range. Both the store parameter and the countby parameter are optional.
See the Query Syntax guide for details on any of these parameters.
query_name = get_facets('()diabetes', facets = 'n.age:range(0,100,10)', store=2)show_hits()
show_hits(hitsDataframe, facetsDataframe=None)
This will display the output into an easy-to-read table when used with get_hits() and get_facets(). It replaces the former response=Query() statement. Show_hits may have two parameters: hitsDataframe and facetsDataframe=None. HitsDataframe is always required; facetsDataframe defaults to None and is not required if charting on the facets is not required.
When writing the query, always replace hitsDataframe with the name of the query used with get_hits().
Below is an example of how show_hits() is used when faceting or charting is not needed.
diabetes = get_hits('()diabetes', fields=CLINICAL_REPORTS_FIELDS)
show_hits(diabetes)To view the query results in list view for an easy-to-read table, include show_hits(hitsDataframe) in the code. Replace hitsDataframe with the variable name assigned to the query.
insomnia = get_hits('(insomnia n.age:[20 TO 60])', fields = 'extract.patient_sex, extract.age, extract.patient_id, extract.patient_full_name, extract.patient_date_of_birth')
show_hits(insomnia)For facets, enter the name of the query as the first parameter and the name of the facet query as the second parameter.
diabetes = get_hits('()diabetes', fields='extract.age')
facet_diabetes = get_facets('()diabetes', facets='n.age:range(0,100,10)', store=2)
show_hits(diabetes, facet_diabetes) #This displays the hits for the query diabetes and allows charts to be made from the facetsUsing get_hits() and get_facets() together can cause the data to be out of sync. This is because the facet query can be written calling different information than the hit query. When this occurs, the person running the report is unable to see the data that is used to create the facets. See the example below:
hits = get_hits('()pickle', fields=CLINICAL_REPORTS_FIELDS, store=0, outputset=True)
facets = get_facets('()earache',facets='d.date_of_service:range(2012-01-01, 2015-01-01, 3month)', store=0)
show_hits(hits, facets)The best way to work around this problem is to write the report with two hit queries along with the facet query: the report segment below contains one that searches for all documents with the term pickle (this will be used in a later part of the report) and another that provides the information that the facets query is based on.
in_a_pickle = get_hits('()pickle', fields=CLINICAL_REPORTS_FIELDS, store=0, outputset=True)
search = '()earache'
store = 3
hits = get_hits(search, fields=CLINICAL_REPORTS_FIELDS, store=store, outputset=True)
facets = get_facets(search, facets='n.age:range(10,16,1)', store=store)
show_hits(hits, facets)
...get_distinct_values()
get_distinct_values(q, fields, store=None)
Use get_distinct_values() to see all unique values in the specified fields. This function will return the distinct values for each of the fields provided. Both the q (query expression) and the fields parameters are required. Multiple fields may be used; as in other queries, list them as a comma separated string.
The function returns a Python dictionary whose keys are the field names (extract.sending_facility) and the corresponding values are Pandas Series objects (BAPTIST MEDICAL CENTER and 7152). The Pandas Series lists all the unique values for the field; the series object's data contains the corresponding record counts.
dv_dict=get_distinct_values('()UNIVERSE', fields='extract.sending_facility, extract.patient_id', store=2)
print(dv_dict)A truncated response to this query is below. It provides the unique value as well as the count of items that have that value. For instance, there are 7,152 records with BAPTIST MEDICAL CENTER SOUTH.
Response:
{'extract.sending_facility': extract.sending_facility
BAPTIST MEDICAL CENTER SOUTH 7152
CALLAHAN EYE FOUNDATION HOSPITAL 7361
...
SHELBY BAPTIST MEDICAL CENTER 6716
SOUTHEAST MEDICAL CENTER 7307
dtype: int64, 'extract.patient_id': extract.patient_id
1141 18
1190 22
...
10336031 1
10523462 1
dtype: int64}Get the number of unique values within a field by using the len method. Below are some options that can be used:
print('Number of Entries: %s' %len(dv_dict['extract.sending_facility']))p_series=dv_dict['extract.sending_facility']
print('Number of Entries: %s' % len(p_series))Sometimes formatting the response is desired. See the script below for an example of one way the information from the query can be used with additional code.
Example:
dv_dict=get_distinct_values('()UNIVERSE', fields='extract.sending_facility,extract.patient_id')
p_series=dv_dict['extract.sending_facility']
print('Number of Entries: %s' % len(p_series)) #Prints "Number of Entries:" and returns the total number of unique values in the facility field.
for tup in p_series.iteritems(): #Iterates over the list and prints out the key (facility name) and the count of records associated with the key
print("%s ....... %s" % tup)Response:
Number of Entries: 17
BAPTIST MEDICAL CENTER SOUTH ....... 7152
CALLAHAN EYE FOUNDATION HOSPITAL ....... 7361
CHEROKEE MEDICAL CENTER ....... 2
CRENSHAW COMMUNITY HOSPITAL ....... 6695
DALE MEDICAL CENTER ....... 6710
DEKALB REGIONAL MEDICAL CENTER ....... 7079
EAST ALABAMA MEDICAL CENTER AND SNF ....... 7423
ELBA GENERAL HOSPITAL ....... 7016
ELIZA COFFEE MEMORIAL HOSPITAL ....... 7086
GEORGE H. LANIER MEMORIAL HOSPITAL ....... 7088
HELEN KELLER MEMORIAL HOSPITAL ....... 7369
JACKSON HOSPITAL & CLINIC INC ....... 7253
MARSHALL MEDICAL CENTER SOUTH ....... 7343
MIZELL MEMORIAL HOSPITAL ....... 6942
SHELBY BAPTIST MEDICAL CENTER ....... 6716
SOUTHEAST MEDICAL CENTER ....... 7307get_unique_values()
get_unique_values(unique, q='()query string', store=None)
Using get_unique_values() provides a list of the unique values within a field. This function runs faster than get_distict_values(), but the end result is similar. However, it is only available for fields that are already prepared for facets.
The unique parameter is the only required parameter and is a string list of field names. More than one field may be used if the fields are separated by a comma, unique='field1, field2'.
The query string is optional and can be used to further restrict the records fulfilling the query. It is important to note that when using the query parameter, q= must be included.
Using store, also comma separated for multiple stores, restricts responses to only the stores provided. Using None or excluding the store parameter, as always, will draw responses from all stores.
To find which fields are available for searching with get_unique_values() use the following code:
find_unique = get_unique_values(unique='""')
print(find_unique)The following query searches for all unique facilities and then returns all facilities that have at least one record with the term Gaea found within it:
Query
gaea=get_unique_values(unique='s.sending_facility', q='()Gaea')
print(gaea)Response
{'s.sending_facility': ['CHEROKEE MEDICAL CENTER', 'MARSHALL MEDICAL CENTER SOUTH']}get_hits_total()
get_hits_total(q, store=None, outputset=None, countby=None)
Using get_hits_total() will run on the query and return the number of records in the search server that match the query criteria. It is important to note that using this function alone returns only a total and will not display hits; use get_hits() and show_hits() if hits are desired.
Query:
diabetics = get_hits('()diabetes', fields = CLINICAL_REPORTS_FIELDS, store = 1)
total_hits = get_hits_total('()diabetes', store = 1)
print(total_hits)Response:
50854get_result_string()
get_result_string(q, fields, store=None, limit=-1, facets=None, format='xml', countby=None)
This function returns the raw string containing the hits from the search server. It replaces the former .raw_execute() function. The seven parameters allowed with get_result_string() are q, field, store, limit, facets, countby, and format. See the Query Syntax guide for details on each of these parameters.
query_name = get_result_string('()UNIVERSE', fields='extract', format='csv')The string size is constrained by the search server to approximately 1 GB.
output_plot()
print(output_plot(clickable=False))
In order to plot a graph, use the helper function output_plot() created by IMAT as seen below. Notice the use of the Pandas .plot() prior to printing the helper function output_plot().
dataframe.plot(kind='bar')
print(output_plot())If more advanced functionality is needed, use the axis object after the dataframe.plot but before print(output_plot()):
ax = dataframe.plot(kind='hist')
ax.set_xlabel('my x label')
ax ... #set labels, titles, legends, ticks, etc.
print(output_plot(clickable=True))Refer to the Pandas Plotting document for more detail on formatting graphs.
Set Functions
Below are some wrapper functions created for the ease of combining and creating sets for viewing and analyzing results. Use these functions both in Report Builder as well as in Set Definitions found in the Admin tab. All set functions must be prefixed by qi. (qi is short for query interface).
- do_set_operation
- gen_set_last_patient_record
- create_set_directly
- update_set
- temp_set_name
- delete_temp_sets
- delete_sets
do_set_operation
qi.do_set_operation(expression)
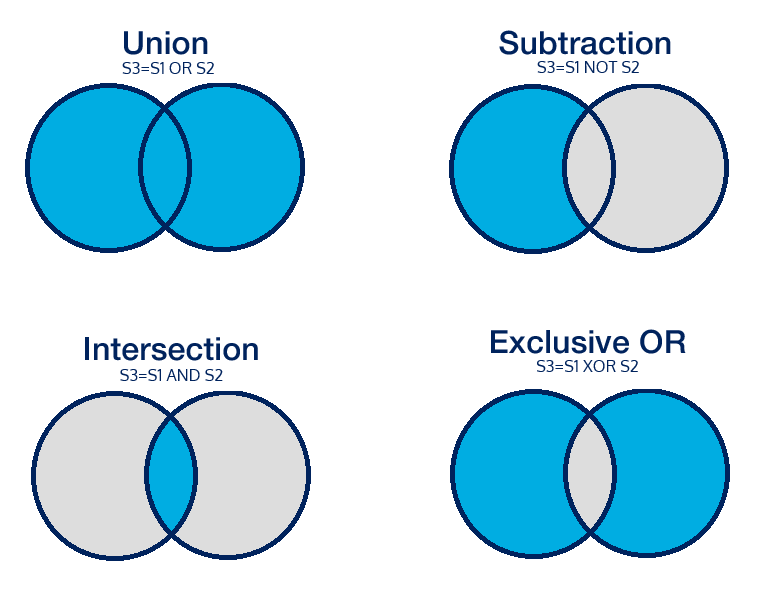
This function combines sets for inclusion and exclusion of data by using Boolean operators. The diagrams on the left illustrate combinations of sets with the blue section highlighting the results for each of the combinations. Set combinations are not limited to two sets and can contain as many sets as necessary. Use any previously created set by entering the name into the function; remember to include the double underscores before and after the set name if using a system set. Note the use of the colon to define the new set.
In the below function, the new set, s5, is a result of a union of s1, s2, s3, and s4.
qi.do_set_operation("s5:s1 OR s2 OR s3 OR s4")Below the four sets (acute_myeloid_patients, chronic_myeloid_patients, acute_lymphocytic_patients, and chronic_lymphocytic_patients) are all previously created and are being combined to make one large set (leukemia_patients).
qi.do_set_operation("leukemia_patients: acute_myeloid_patients OR chronic_myeloid_patients OR acute_lymphocytic_patients OR chronic_lymphocytic_patients")However, if you were to create an intersection of the four sets, you'd get a smaller result list because the results must contain only hits that meet all four criteria.
qi.do_set_operation("s5:s1 AND s2 AND s3 AND s4")Combine Boolean operators to create an accumulation of sets. However, note that qi.do_set_operation does not support internal parentheses and the operations are always applied left to right regardless of the usual order of operations.
qi.do_set_operation("s5:s1 OR s2 NOT s3 AND s4")The above example is the equivalent of the following:
sa=s1 OR s2
sa=sa NOT s3
s5=sa AND s4gen_set_last_patient_record
qi.gen_set_last_patient_record(q, store, outputset)
Note: All three parameters must be entered, and they must be entered in this order: q, store, outputset.
To generate a list of the last patient records loaded into the system, use this function. Use any query expression desired following the query syntax rules. If the query expression is empty, then UNIVERSE is used by default.
Only one store can be used at a time. A store name can be entered instead of a number, just remember that all string inputs need to be surrounded by quotes.
Enter the name of the outputset. This name is a previously created set.
Below the example shows the previously created system set named __itchyPeople__ that includes all people with asthma, allergies or eczema. The user now is searching for the last patient record within the last year for each patient.
qi.gen_set_last_patient_record("d.date_of_service:[-1y TO TODAY]", 1, "__itchyPeople__")create_set_directly
qi.create_set_directly(outputset, jsonbody)
You may find that you want to create a set by using your own array of information rather than relying on a query. To create a set directly, use outputset to define the new set's name and JSON syntax to create the array for jsonbody. See the example below for the proper way to create a set with known values from the patientIDN field. Notice that MPID_json is the jsonbody parameter listed above and contains my own array of MPIDs, and that the field name is included in parentheses after the new outputset name of mpidsIWant.
MPID_json="['mpid1', 'mpid52', 'mpid54', 'mpid55', 'mpid112']"
outputset="mpidsIWant(patientIDN)"
qi.create_set_directly(outputset, MPID_json)
Alternatively, you can set the outputset parameter in the wrapper function instead of setting it on a separate line:
qi.create_set_directly("mpidsIWant(patientIDN)", MPID_json)
It is also possible to create new sets based on spaces and record reference numbers. Space numbers correspond with store numbers. Once again, proper JSON syntax is needed to create the JSON body. You must use the word space before the corresponding number. After the space has been set, include an array of record references:
jsonWithSpaces={ "space0" : [r1, r2, r3],
"space2" : [r1, r5, r10],
"space5" : [r1, r2, r22]
}
outputset="spaces"
qi.create_set_directly(outputset, jsonWithSpaces)
Alternatively, you can set the outputset parameter in the wrapper function instead of setting it on a separate line:
qi.create_set_directly("spaces", jsonWithSpaces)
update_set
qi.update_set(q, outputset, store=None)
If a previously created set is out of date because more records have been fed into the system, you can update the set by using qi.update_set(). It is faster to update the set than to remake the set using all the records found in the system. This is because updating only requires a query of newly fed documents rather than the entire corpus of data. This function uses the time stamp of the previously created set and creates a union from the previously created time to the present.
If a set does not exist, running an update creates the set.
qi.update_set('()obese diabetes', outputset='obese_diabetic_patients(patientidn)') #<em>obese diabetes</em> is the base query for the original set; <em>obese_diabetic_patients(patientidn)</em> is the name of the original set; you can also add a store or stores here
df=get_hits('()FILTER(patientidn:SET:obese_diabetic_patients)', fields=CLINICAL_REPORTS_FIELDS)
print(df)#optionaltemp_set_name
qi.temp_set_name(prefix, fields = None)
To define a temporary set, use temp_set_name. The function qi.temp_set_name returns a tuple with the tmpSetName and tmpOutputSetName. A comma to separates the tuple pieces. You must use qi. to prefix the function. Include both the prefix name (you determine this, it is not found anywhere else in the data) and the quick table field you are choosing to search. The field is not required if you are doing a record-centric search rather than a patient-centric search.
tmpSetName, tmpOutputSetName = qi.temp_set_name('myprefix', 'patientidn')Second, create your set using outputset and the variable previously chosen. The standard sets syntax applies.
get_hits_total(q="my query string to create set", outputset=tmpOutputSetName)Third, write your query using .format and the tmpSetName. At this point, all query parameters and Boolean syntax can be used.
df = get_hits(q="()FILTER(patientidn:SET:{})".format(tmpSetName), fields="field1, field2")The example shown below uses a quick table field. If you choose to use a NREF set instead, the steps are the same, but do not specify a field parameter in the qi.temp_set_name function.
Example:
diabetics, outdiabetics=qi.temp_set_name('diabet', field='patientidn')
print(diabetics) #optional
print(outdiabetics) #optional
patient_count=get_hits_total('()diabetic', outputset=outdiabetics, countby='patientidn')
print(patient_count) #optional
#dataframe of clinical records for the patients
df=get_hits("()metformin FILTER(patientidn:SET:{})".format(diabetics), fields=CLINICAL_REPORTS_FIELDS)
print(df)
Response
tmp.diabet.1570134770.0931187
tmp.diabet.1570134770.0931187(patientidn)
9999
uri ... extract.record_type
0 Jonash-20-osteotrans-ORUR01/ARIA-0000034513.hl7 ... transcription
1 Jonash-7-trans-ORUR01/DAREN-0000011761.hl7 ... transcription
2 Jonash-20-osteotrans-ORUR01/WILBUR-0000030847.hl7 ... transcription...
delete_temp_sets
qi.delete_temp_sets(deleteList)
Under normal circumstances, there is no need to call this function. The search server is configured to delete all temporary sets after the compound query finishes. However, if you are using multiprocessing, this function is needed to remove temporary sets. It should appear in the finally block of the multiprocessing proc function.
def Proc(...): # multiprocessing proc function
try:
# Code that calls temp_set_name(...)
finally:
qi.delete_temp_sets()delete_sets
qi.delete_sets(deleteList)
This function rarely needs to be called because the compound query runner always calls delete_temp_sets() as it exits the main processing thread. However, if you wish to explicitly delete non-temporary sets, you can do so. However, once a set is deleted, it is permanently removed and you cannot reverse that delete. You'll have to create the set again if you find you need the set.
Because you are including a list of sets to delete (even if there is only one set), you must enclose it in brackets, [ ], like any Python list.
qi.delete_sets(['setName1', 'setName2', 'setName3'])CLINICAL_REPORTS_FIELDS
When using the IMAT product, use the shortcut fields=CLINICAL_REPORTS_FIELDS instead of individually entering the below fields. This pulls all fields mentioned below and display them in the list of Hits on the IMAT Solutions product. If CLINICAL_REPORTS_FIELDS is not used and the fields are not entered individually, some columns on the Hits table will display Unavailable.
- uri
- extract.patient_id
- extract.patient_full_name
- extract.patient_date_of_birth
- extract.date_of_service
- extract.sending_facility
- extract.record_type
- store
Any fields aside from those above can be entered if you add them to the query with a plus sign, +, and include both an initial comma and the fields in quotes. Include extract. in front of the field name if the field comes from the extract.
hits = get_hits('()heart disease', fields=CLINICAL_REPORTS_FIELDS + ',extract.age', store=2, limit=25)
show_hits(hits)Layout Decisions
The layout of compound queries is a loose overall structure and consists of three parts: setup, processing, and output. It is unnecessary to strictly adhere to this exact layout, but it is best practice to enter the data in this order.
Setup
The setup consists of assigning variables, setting up imports and helper functions, and creating queries using the information on the search server.
It is possible to write lists, sets, and dictionaries in a vertical list with each item on its own line. A vertical layout proves easier to read when the list contains many items. When compiling the list in this manner, the items in the list still need to be separated by a comma. Indent all lines except the first; if formatted correctly, the script should automatically indent. The final line should be the closing punctuation mark: bracket, bracket-parenthesis, or brace.
myHospitals = ['Mountain Valley Hospital',
'Urban Urgent Care',
'Suburban Hospital',
'Rural Hospital',
'County Radiology',
'Dialysis Center'
]Creating a query in a compound report script refers to pulling information from the search server. To format the query correctly, use the following format:
query_name = get_hits('(Boolean Expression)', parameter1 = 'value', parameter2 = 'value')If using a field name as part of a Boolean expression, enter the terms in the Query Builder section of a single report to see how it should be formatted: age becomes n.age:[25 TO 35]. When using field names as a parameter of a query, indicate where the field is located; two examples include the quick table and the extract:
query_name = get_hits('(diabetes n.age:[25 TO 35])', fields = 'qtable.patient_id, extract.age', limit = 10)Processing
The processing portion refers to calculating data acquired in the setup segment of the compound query. It is in this section that counts, averages and other mathematical functions are processed. It is also this section where data is aggregated from various stores, and the commands or functions for data is written.
Output
The final part of a compound query is the output section. Here it is determined where the results of the query are sent. It can be sent to a file or to the output pane of the report builder. The output pane of the Clinical Reports page supports HTML and has limited support for JavaScript.
Non-Results Output
Output from compound queries is normally returned using the same format used in simple queries. Usually this is a set of results computed from a query, or aggregated from multiple queries. When running a compound query, it is often the case that the items returned from the queries are much less important than the analysis run on those items. To return non-result information from the query, use the print function to the stdout tag of the response.
All output to standard out and standard error are captured and returned under the <stdout> and <stderr> tags.
Server Response
In addition to the normal tags returned by a simple query, the following tags are added to the XML:
- result - Contains the exception or error information to be displayed in cases where no results are returned.
- stdout - Contains all information written to stdout when the query is executed.
- stderr - Contains all information written to stderr when the query is executed.
- python_query - This tag contains the entire Python code executed on the server. This code is formatted for display by the browser.
- traceback - This tag contains the Python trace stack formatted for display in the browser. The line numbers should match the code provided under python_query.
Maximizing Performance
While the search appliance is tuned to provide the best performance available, compound query exposes functionality that allows the user to perform tasks in ways that are less-optimal. The following guidelines will help keep the system performing as expected to make the most of the search appliance.
Use the Smallest Sets Possible
When running queries to get data for analysis, provide as many constraints to that set as possible. Returning items from a query and then testing them to see if they meet the criteria is much slower than using the search index to eliminate them from the returned results.
Avoid Disk IO
When running queries, use the fields parameter to return only the needed data. Reducing the amount of data returned, more information will fit into memory, increasing performance. It is also helpful to tune the parse table, moving frequently used data elements into the extract.